May 7, 2024 — A new study by researchers from the Hebrew University of Jerusalem sheds new light on the intricate process of mRNA regulation during embryonic development, providing novel insights into how pluripotent cells adopt specialized identities through gene expression.
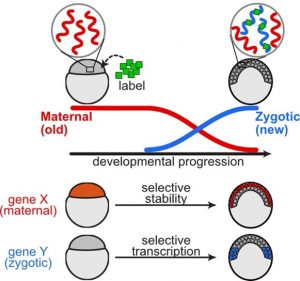
Understanding mRNA regulation during embryonic development helps decipher how genes are turned on and off in specific cells at precise times. Embryonic development involves pluripotent cells assuming specialized identities by adopting particular gene expression profiles. However, understanding the relative contributions of mRNA transcription and degradation to shape these profiles has been challenging, particularly within embryos with diverse cellular identities.
The study, published in Nature Communications, combines single-cell RNA-Seq and metabolic labeling in zebrafish embryos, distinguishing between newly transcribed and pre-existing mRNA. This approach quantifies mRNA transcription and degradation rates within individual cell types, uncovering varied regulatory rates across genes and cell-type-specific differences in degradation.
The findings reveal highly varied regulatory rates across thousands of genes. The researchers observed coordinated transcription and destruction rates for many transcripts and linked differences in degradation to specific sequence elements. Importantly, they identified cell-type-specific differences in degradation, including selective retention of maternal transcripts within primordial germ cells and enveloping layer cells, two of the earliest specified cell types.
The research was led by Ph.D. student Lior Fishman and a team under the guidance of researcher Dr. Michal Rabani at the Alexander Silberman Institute of Life Science at Hebrew University, in collaboration with researchers from the U.S. National Institutes of Health.
In the study, researchers used a technique called single-cell RNA sequencing along with metabolic labeling to track how genes are turned on and off over time in zebrafish embryos. They could tell apart the mRNA that was made new (from the embryo itself) and the mRNA that was already there (from the mother). Using mathematical models, they measured how fast genes were turned on and off in different types of cells as they developed.
Dr. Rabani said, “Our study provides a quantitative approach to studying mRNA regulation during a dynamic spatio-temporal response. This work opens up new avenues for understanding the molecular mechanisms underlying cell fate determination during embryonic development. We hope this will pave the way for future studies aimed at unraveling the complexities of gene expression regulation in various biological contexts.”
The research paper titled “Cell-type-specific mRNA transcription and degradation kinetics in zebrafish embryogenesis from metabolically labeled single-cell RNA-seq” can be accessed at https://doi.org/10.1038/s41467-024-47290-9.